CarrierCheck: End-to-end Solution to Inherited Disease Screening from Wet-Lab to Bioinformatics
Funding : Eureka Secretariat
Program: Eureka Network Projects
Consortium Partners: Genoox (Israel)
Project Budget: 2.3M €
Period: 2022-2024
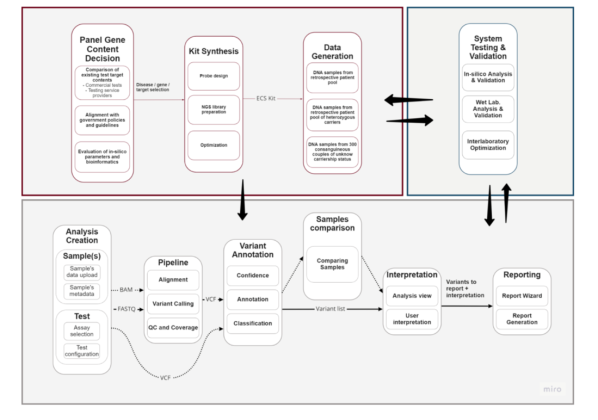
Scope of the Project: Within the scope of our project, we plan to produce a carrier screening test with the commercial name “CarrierCheck” and a kit that can be used by genetic diagnosis centers for test service purposes. CarrierCheck promises to be a screening test with high analytical efficiency and clinical validity, covering the entire testing process from end to end. It has been developed for diseases of significance in European and Middle Eastern populations, with unique genetic content. The test is designed for quick application, suitable for detecting challenging regions through Next-Generation Sequencing (NGS), guaranteeing high-quality sequencing, adaptable to widely used NGS platforms, and usable by clinicians from various fields. This method, which enables the analysis and reporting of all variants causing the disease with a single NGS kit, represents a first in the carrier screening test market, ensuring more reliable reproductive decisions.
The aim of the project is to develop a pan-ethnic and highly clinically useful commercial carrier screening test for hereditary rare diseases for use in reproductive medicine without the need to know the ethnicity and family history of the individual.
The test consists of three components: a genetic screening panel, an integrated analysis software and an interpretation and reporting interface. Each community needs different approaches to large multigene screening tests -ECS – that are compatible with its epidemiology.
In this project, gene and target region curation will be carried out by considering the content of existing commercial panels and Mikrogen’s 20 years of experience in Turkey and the Middle East. Wet lab and bioinformatics processes will be optimized specifically for each targeted disease/gene/variant, considering the bioinformatics challenges arising from the nature of wet lab and next generation sequencing (NGS) data, in order to obtain results with high sensitivity and specificity. Genoox will assess the genomic characteristics and data analysis needs of each targeted region and develop additional bioinformatics solutions alongside traditional analysis algorithms for challenging regions. The final output of the project will be a pan-ethnic and clinically useful carrier screening test for use in reproductive counseling.
CarrierCheck: End-to-end Solution to Inherited Disease Screening from Wet-Lab to Bioinformatics
Funding Institution: TÜBİTAK (Scientific and Technological Research Council of Turkey)
Program: 1501 / Industrial R&D Projects Support Program
Consortium Partners: PhiTech (Türkiye)
Project Budget: 5M ₺
Period: 2021-2023
Scope of the Project: The diagnosis rate of rare genetic diseases remains at the level of 50% with DNA-based sequencing analyzes routinely used in the clinic. The low number of cases encountered in the population and the high phenotypic and genotypic heterogeneity of these cases cause limitations in the diagnosis of rare diseases and thus in the management of these diseases. RNA-Seq data constitute a very important resource as a solution to the problem of inadequacy of routine genomic analysis in diagnosis. With these data, not only transcriptome information but also additional information such as variant information and splicing (end joining regions) information can be obtained. In the literature, it is reported that the inclusion of RNA-Seq data in analyses contributes to diagnostic power. Within the scope of the project, it is aimed to increase the diagnosis rate in rare diseases thanks to a workflow that integrates multi-omics approaches and patient phenotype information.
Next Generation Sequencing (NGS) technologies enable the detection of disease-causing variants in a short time and cost-effectively. With the introduction of NGS technology in the clinic, the capacity to identify genetic disorders in both known and novel disease genes has increased. Following the discovery of 555 genes associated with Mendelian diseases using NGS between 2010 and 2015, Whole Exome Sequencing has become the current method used for the diagnosis of heterogeneous rare diseases with suspected Mendelian inheritance. Although WES technology, which is defined as the sequencing of protein-coding parts constituting 1-2% of the whole genome, is routinely used in the clinic, it enables diagnosis in only 30%-50% of rare mendelian diseases depending on the type of disease and patient selection (Fresard and Montgomery, 2018). Many factors can be effective in achieving these results in the clinic. These includes;
- i) Presence of a large number of variants of unknown significance (VUS) as a result of exome sequencing and the inability to interpret these variants due to their unknown association with diseases,
- ii) İnability to analyze structural variants, intergenic regions, triple repeats, mitochondrial gene regions with WES,
iii) Complex inheritance problem.
For these reasons, in order to diagnose rare genetic diseases, there is a need for additional information such as gene expression level, abnormal end joining analysis, which cannot be provided by WES but can be obtained from RNA seq data. In many studies in the literature, it has been shown that the diagnosis rate, which is 25-50% using WGS and WES data, increases by 10-35% with the contribution of RNA-Seq data. Although studies have shown the effect of RNA Seq data on diagnosis, RNA Seq data is not yet used as a diagnostic method in clinical applications.
Within the scope of the project, it is aimed to increase the diagnosis rate of rare diseases, which remains at 50%, and to diagnose patients who cannot be diagnosed with WES in the light of information obtained from RNA seq data. For this purpose, a best practice workflow that can be used routinely in the clinic and improved for rare diseases will be developed.
For this purpose, standards for both sequencing and analysis will be determined by PhiTech and Mikrogen for RNA-Seq to enter clinical practice. Reproducibility and quality data acquisition in sequencing will be ensured by Mikrogen, while the part related to bioinformatics analysis will be developed and optimized by PhiTech. The creation of the candidate and causal variant list to be obtained as a result of the workflow developed within the scope of the project will be provided by the cooperation of the two companies.
CarrierCheck: End-to-end Solution to Inherited Disease Screening from Wet-Lab to Bioinformatics
Funding Institution: Ankara Development Agency
Program: Medical Devices Financing Support Program
Project Budget: 2M ₺
Period: 2021
Scope of the Project: Current carrier and screening kits used in the market are produced by international biotechnology companies. However, these kits do not cover all genes associated with genetic diseases frequently encountered in our country. With the screening kit based on next-generation sequencing technology to be developed within the scope of the project, mutations that cause genetic diseases commonly encountered in the Turkish population will be identified before marriage. In this context, our team’s more than 20 years of experience in the field of medical genetics with more than 600,000 patient samples and state-of-the-art equipment park constitute an important infrastructure for the study. The screening panel kit developed within the scope of the project was adapted to the recently developed MGI sequencing device, which enables low-cost sequencing of a large number of samples. In this way, it was possible to design a kit targeting more rare diseases instead of only a limited number of common single gene diseases, and it was also possible to screen for genetic diseases, especially those caused by inbreeding. This low-cost broad screening panel kit is a test that can be used in domestic and international screening programs, especially in public health.
CarrierCheck: End-to-end Solution to Inherited Disease Screening from Wet-Lab to Bioinformatics
Funding Institution: TÜBİTAK (Scientific and Technological Research Council of Turkey)
Program: 1505 / University-Industry Cooperation Support Program
Project Budget: 1M ₺
Period: 2019-2021
Scope of the Project: The need for in vitro fertilization (IVF) procedures is increasing day by day both globally and in our country. However, the success rate of IVF procedures falls below the desired levels. One of the fundamental reasons for this situation is the incorrect timing of embryo transfer to the endometrium. Among the main deficiencies of the kits available in the market are their use of solid biopsies, their inability to reflect real-time test results, and their inability to be used for ongoing IVF treatments. In response to these issues, this project aims to detect and predict the time window known as the “implantation window” from blood and to enhance IVF success rates through minimally invasive interventions for IVF patients.
The method to be employed in increasing the IVF success rate involves the detection of miRNA-type biomarkers. There are findings in the literature that suggest the existence of miRNAs that could assist in determining the correct implantation day (IP). However, such miRNAs need to be detected in endometrial tissue. Unlike kits that achieve this, within the scope of this project, a superior kit has been developed using a minimally invasive approach that can provide information for the same menstrual cycle.
For this purpose, discovery and validation studies of microRNAs (miRNAs) capable of determining the most suitable day for embryo implantation in the blood have been conducted. The discovery of miRNAs was carried out using in-silico methods and RNA sequencing. The conducted miRNA examinations were used both for detecting level differences and for identifying RNA editing events present in miRNAs.